The Hamza Lab is a world leader in the study of heme, a vital but cytotoxic cofactor essential for life. Our question is simple: How do cells and organs coordinate heme availability to sustain life? We move beyond the textbook view of isolated cells to explore cell-nonautonomous regulation, where organs and organelles communicate through sophisticated heme-signaling networks to maintain systemic health.
RESEARCH SUMMARY
The Global Impact: Heme as a Vital Nutritional and Clinical Axis.
Iron deficiency remains the world’s most prevalent nutritional disorder, with nearly 80% of the population affected. While inorganic iron is difficult to absorb, heme is a highly bioavailable iron source that bypasses many dietary barriers. Beyond nutrition, our bodies recycle five million red blood cells every second, a massive daily iron-recycling effort mediated by macrophages. Our lab bridges public health and clinical medicine by identifying the elusive molecular pathways and transporters, such as HRG1, that govern how this vital cofactor is absorbed, recycled, and utilized.
The Mechanistic Challenge: Moving a Toxic Necessity.
Heme is a double-edged sword: it is essential for physiological processes ranging from gas sensing to microRNA processing, yet its hydrophobic and reactive nature makes free heme highly cytotoxic. Despite its importance, the pathways that safely traffic heme through membranes to target organelles remain poorly understood. We challenge the traditional cell-autonomous view of heme regulation by exploring how complex intercellular trafficking networks and inter-organ signaling coordinate systemic homeostasis to prevent oxidative damage while sustaining essential metabolism.
Our Approach: Innovative Models and Paradigm-Shifting Discoveries.
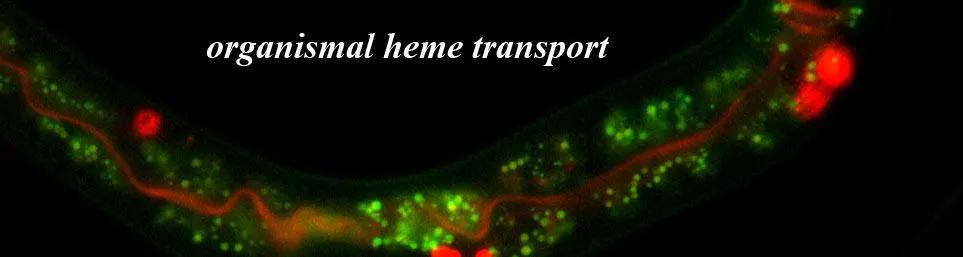
To solve these mysteries, we employ a diverse toolbox of genetically-tractable models, most notably the microscopic transparent animal Caenorhabditis elegans. As a heme auxotroph that cannot synthesize its own heme but needs heme to survive, this roundworm provides a clean genetic background to pinpoint human orthologs and define the first molecular frameworks for heme signaling. By integrating these findings with other sophisticated model systems including genetically-engineered mice, zebrafish, yeast, Leishmania, and human organoids, we are redefining our understanding of iron metabolism, host-pathogen competition, and the fundamental mechanisms governing organismal lifespan
Why join our team?
Our lab provides a unique, interdisciplinary environment where you will lead high-impact projects that bridge the gap between basic discovery and human disease. We are a collaborative group, frequently integrating efforts with research groups around the world, to combine biophysical, biochemical, and molecular genetic approaches that no other group can replicate. We seek curious and driven researchers eager to discover how this essential cofactor shapes organismal homeostasis. We invite you to explore our publications and reach-out to discuss how your skills and ideas might help drive our next breakthrough.
- Advanced Genetic Modeling: Work with a diverse array of model systems, from the heme-auxotroph C. elegans, to yeast, CRISPR-engineered zebrafish to mice, and infectious Leishmania pathogens.
- Cutting-Edge Omics: Gain expertise in single-cell RNA sequencing (scRNA-seq), whole-organ RNA-seq, multidimensional protein identification technology (MuDPIT) to map the heme interactome, and lipidomics.
- In Vivo Imaging: Utilize tissue-restricted, genetically-encoded heme sensors and novel spectroscopic imaging systems to visualize real-time heme signaling at subcellular resolution.
- Translational Physiology and Drug Discovery: Investigate clinical challenges including iron deficiency, porphyria, infectious diseases, and the fundamental mechanisms of aging.
MAJOR RESEARCH PROJECTS:
1. Inter-organ Heme Signaling
The Big Picture: For decades, the biological paradigm stated that cells were self-sufficient islands that managed their own heme needs. Our lab is shattering this concept by proving that cellular heme levels are maintained by an inter-organ communication network. Because heme is both a vital nutrient and cytotoxic, organisms must signal their systemic requirements across tissues to prevent deficiency or toxicity.
The Challenge: We utilize the unique heme-auxotroph C. elegans to map these cell-nonautonomous pathways, as they allow us to manipulate body heme solely through diet. We have already identified HRG-7, a protein that communicates heme status between the intestine and neurons, the so-called gut-brain axis. Our current mission is to use forward genetics and proteomics to identify the molecular senders and receivers of these signals.
By joining this effort, you will help us define the very first molecular framework for metazoan heme signaling and show how remote organs coordinate systemic homeostasis, a process essential for understanding mammalian health.
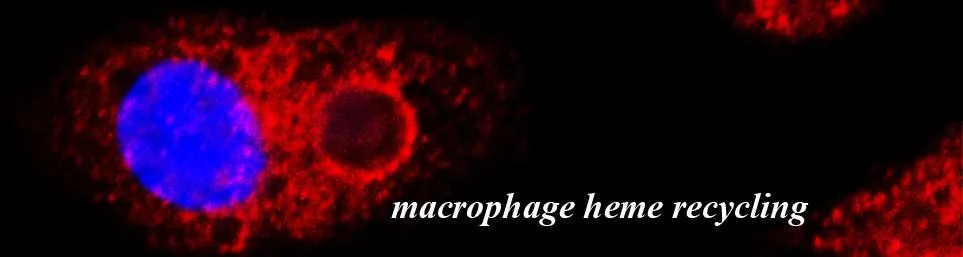
2. Heme Transport and Iron Recycling
The Big Picture: Approximately 90% of our daily iron needs are met by macrophages recycling heme from old red blood cells. Despite its importance, the molecular machinery that moves toxic heme out of the macrophage's phagolysosome ("garbage disposal") remained a mystery for years.
The Challenge: Our lab identified HRG1/SLC48A1, the first and only bona fide eukaryotic heme transporter. While HRG1 is essential for iron recycling, our recent work revealed an unexpected mammalian survival mechanism: when HRG1 is missing, mice can detoxify heme by crystallizing it into hemozoin, a feat previously thought to be exclusive to blood-feeding parasites like malaria. We are now investigating the relationship between HRG1 and other proteins in the pathway to understand how cells make hemozoin and whether cells normally make hemozoin as a heme tolerance system.
By joining this effort, you will help us redefine how the body manages its most precious metal using CRISPR-tagging, MuDPIT proteomics, lipidomics, metabolomics, and RNA-seq to uncover the heme-recycling interactome and mammalian hemozoin pathway.
3. Heme Transport in Hemoglobinopathies and Erythropoiesis
The Big Picture: Hemoglobinopathies, such as beta-thalassemia, affect 7% of the global population and are characterized by a profound imbalance between heme and globin production. For example, in beta-thalassemia, the lack of beta-globin leads to the accumulation of toxic free heme and alpha-globin, causing oxidative stress and the death of erythroid progenitors. While textbooks focus on internal heme synthesis, our lab is challenging this dogma by investigating whether red blood cells also rely on external heme import to coordinate hemoglobin assembly.
The Challenge: We are exploring the role of HRG1 heme importer in the developing red blood cell. Our data show that without HRG1, erythroid maturation is delayed, especially during stress anemia. Because free heme is toxic in beta-thalassemia, we showed that genetically inhibiting HRG1 actually reduced disease severity by reducing excess heme. We are currently using CRISPR-engineered mouse models to determine if targeting this transport pathway can alleviate chronic anemia and iron overload.
By joining this effort, you will help determine if heme transport is a viable new therapeutic strategy for genetic blood disorders by using molecular genetics, flow cytometry, and hematology to redefine the molecular framework of erythropoiesis.
4. Heme Transport at the Leishmania-Host Interface
The Big Picture: Neglected Tropical Diseases like Leishmaniasis affect hundreds of millions worldwide, yet therapeutic options remain limited and toxic. The Leishmania parasite is a master of survival, replicating inside the very cells designed to kill it: macrophages. Our research has uncovered a critical Achilles' heel in the parasite’s biology - it is a heme auxotroph, meaning it cannot synthesize its own heme and must hijack it from the host to survive and reproduce.
The Challenge: We discovered LHR1 (Leishmania HRG1), the first parasite heme transporter, that Leishmania uses to scavenge host heme. Because LHR1 is essential for the parasite, and notably different from human transporters, it represents a perfect target for drug development. We are currently refining a promising lead small molecule that selectively blocks LHR1 to starve the parasite of heme without harming the host cell. Beyond drug discovery, we are investigating the tug-of-war for heme between the parasite and the macrophage, meaning how parasite LHR1 competes with host HRG1 to impact infection.
By joining this effort, you will help bridge fundamental biology with drug discovery by using high-throughput screening, medicinal chemistry, and innovative in vivo imaging to visualize the competition for heme between the host and pathogen.
5. Mammalian Models of Intestinal Heme Absorption
The Big Picture: Heme iron is the most bioavailable form of iron in our diet, yet the mechanism for how our gut absorbs it remains a black box. Inadequate absorption leads to anemia, while excessive absorption causes iron overload. We are establishing the molecular blueprint for this essential nutritional process.
The Challenge: We are investigating whether HRG1 acts as the intestinal heme importer and how the body balances heme vs. iron (non-heme) absorption to maintain balance. We and our collaborators have established genetically-altered mice and rats as models for heme absorption. Our approach spans from in vivo models of disease to human intestinal organoids.
By joining this effort, you will help us clarify the ambiguities of dietary iron uptake and improve treatments for global iron deficiency using genetically-altered vertebrates models and in vivo metabolic modeling.
6. Heme and Organismal Lifespan
The Big Picture: Traditional aging research focuses on mitochondrial decline. Our lab’s exciting new findings are paradigm-shifting: cellular heme can dramatically alter lifespan. We hypothesize that heme acts as a master signaling molecule that coordinates adaptive metabolic responses to aging.
The Challenge: We and our collaborators utilize genetically-encoded heme sensors to monitor labile heme in real-time within specific tissues and organelles. We are investigating how heme signals are transmitted through conduits at membrane contact sites and deploying hemoproteomics to identify the novel targets that control healthspan.
By joining this effort, you will help pioneer the first comprehensive study on how a simple cofactor can govern the pace of life using in vivo genetic reporters, transcriptomics, and genetic mutants.
7. Hepatocyte-Macrophage Crosstalk in Porphyrias
The Big Picture: Medical textbooks suggest the liver manages its heme primarily through internal synthesis. However, we and our collaborators discovered that even when liver synthesis is completely blocked, hepatocytes stay healthy, meaning heme can be acquired from neighboring cells. This unexpected intercellular pathway represents a new target for treating metabolic disorders like Porphyria.
The Challenge: We are mapping the molecular bridge between these cell types. Our research focuses on identifying the transporters and signaling molecules that drive this acquisition process. We utilize single-cell RNA sequencing (scRNA-seq) and advanced proteomics to determine if this system is constitutively active or a conditional emergency response to heme deficiency.
By joining this effort, you will help us uncover the missing links in intercellular crosstalk and metabolic flexibility using both in vitro and in vivo mammalian models.
Our research is supported by grants from the National Institutes of Health.